
Burkina Faso
The ODIN team in Burkina Faso collected pilot samples in October and November 2024. Wastewater, drinking water, soil and seafood (fish and prawn) have been collected for work package 2 and 3 (WP 2 and WP 3).
Water samples for WP3 have been filtered, and the filters as well as soil and seafood were sent to NORCE (Norway) for metagenomics. Preliminary results are available, and data analysis is still in progress.
Samples for WP2, mainly drinking water, surface water and wastewater have been analyzed at Clinical Research Unit of Nanoro (CRUN) microbiology laboratory. Water samples were filtered or diluted and plated on selective culture media according to ODIN laboratory handbook. The priority pathogens selected by ODIN consortium have been looked for, such as Salmonella Typhi, Vibrio cholerae, fecal indicator bacteria (FIB) such as Enterococci and Escherichia coli and antimicrobial resistant bacteria such as extended spectrum beta-lactamase (ESBL) and carbapenemase (CP) producing E. coli and Klebsiella pneumoniae.
Pilot sampling in DRC
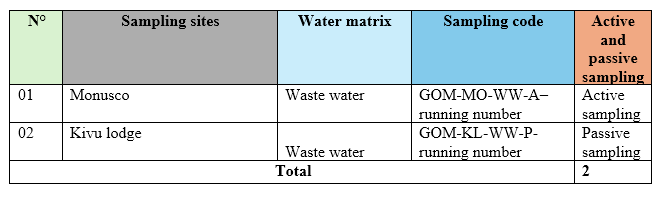
On Tuesday, November 20, 2024, samples were collected from two of the ten selected sampling sites in Goma (East Democratic Republic of Congo), as shown in the table below. The selected sites were MONUSCO for active sampling and Kivu Lodge for passive sampling.
Samples collected on-site in 50ml plastic bottles were labeled with a unique code and stored in an isothermal box at a temperature between 3.6 and 3.3°C (temperature compliant with handbook instructions:5±3°C).
At the INRB microbiology laboratory, upon sample reception, details (sample type, code, date, time, and arrival temperature) were recorded, and the sample reception form was completed.

After enrichment, samples were incubated for 24 hours at 37°C. Cultivation began the following day (Day 2), following the ODIN microbiology handbook procedures and the standardized procedures of the INRB microbiology laboratory. The aim was to isolate the bacteria of interest for the ODIN project: Vibrio cholerae, Salmonella Typhi, Escherichia coli, Klebsiella pneumoniae, and Enterococci. Five culture media were prepared: Blood Agar, MacConkey Agar, SS Agar, Hektoen Enteric Agar, and TCBS Agar.
Pilot sampling in Tanzania
Pilot sampling took place in two sites, Tanga and Dar
On 21 October 2024, four samples were collected from two locations in Tanga, as shown in the table below. Furthermore, the ODK collect App was used to complete and upload sample collection details using the ODIN Field Sample Collection Form.

Following the pilot sampling, regular sampling and analysis have commenced across the three sites. Update on this will be provided shortly.
Culture-based microbiological analysis of water samples
When received at the NIMR Tanga microbiology laboratory, all four sample details, including sample type, code, date, time, and arrival temperature, were recorded and uploaded into the ODK collect App by completing the ODIN Sample Receiving Form-Tanga.
The two water-based samples were analyzed using the ODIN Microbiology Handbook to isolate priority organisms, i.e., Vibrio cholerae, Salmonella Typhi, ESBL-producing Escherichia coli, and Klebsiella pneumoniae,Carbapenemase-producing Enterobacterales (CPE) such as Escherichia coli and Klebsiella pneumoniaeand Feacal Indicator Bacteria (FIB). The two soil samples were stored (waiting for a standardized procedure). In addition, a fraction of 100mL for each raw sample was stored for later molecular analysis.
For two consecutive days (23 and 24 October 2024), a total of 14 samples were collected from nine locations in Dar es Salaam as shown in the table below. Furthermore, the use of ODK collect App was applied by completing and uploading sample collection details using ODIN Field Sample collection Form.

Also By
- 1er webinaire ODIN : le consortium partage ses connaissances sur les stratégies d'innovation, de résilience et de durabilité
- 1st ODIN Webinar: The Consortium Shares Insights on Innovation, Resilience, and Sustainability Strategies
- Projet ODIN de Surveillance des eaux usées : Campagne d'échantillonnage terminée
Please Sign in (or Register) to view further.